PaPIM-CP2K_Interface¶
Purpose of Module¶
Module PaPIM-CP2K_Interface couples the PaPIM code with the CP2K program package, where the latter is used for calculation of system electronic structure properties. It directly links CP2K as a library for potential energy calculations to the PaPIM code and avoids the significantly slower exchange of information between the two codes by reading and writing to an external file. The CP2K program package provides a general framework for different modeling methods such as DFT using the mixed Gaussian and plane waves approaches, semi-empirical methods and classical force fields. This enables virtually any calculation of time-dependent correlation functions for any system, without depending on the availability of analytical potentials for the studied system, as was the previous case in PaPIM code. Using the MPI split communicator approach the CP2K subroutines can be executed on multiple cores for each sampling trajectory, enabling a parallel calculation of system potential energy and gradient values.
Applications of the Module¶
The inclusion of CP2K for computation of system’s electronic structure properties enables calculation of time-dependent correlation functions to a vast range of systems, while CP2K can perform atomistic simulations of solid state, liquid, molecular, periodic, material, crystal, and biological systems. The PaPIM code has also been upgraded with periodic boundary conditions to enable simulations of solid and liquid state systems. For any system whose properties can be determined with the CP2K code, a corresponding time-dependent correlation function can be computed now with the PaPIM code.
Compiling¶
In order to compile this module the CP2K program package has to be properly set-up and the CP2K has to be
compiled as a library as well.
In the latter case, the CP2K root directory contains a sub-directory lib which
contains the corresponding library files.
In the absence of the latter, CP2K cannot be linked to PaPIM code.
For information on installing the CP2K code and compiling it as a library the user is advised to examine the
CP2K installation documentation at this link.
Fortran compiler with a MPI wrapper together with lapack libraries have to be available to successfully
compile the code.
The user is advised to examine the Makefile in the ./source sub-directory prior to code compilation
in order to select an appropriate compiler and to check or adapt the compiler’s options
to his local environment, or to generally modify the compiler options to his requirements.
Special care should be made on the CP2K paths to the corresponding library files on certain systems.
The Makefile contains two example cases encountered on cluster systems (which use Intel compilers)
but any other variation is possible.
The compilation flag --D__USE_CP2K controls the inclusion of CP2K into the PaPIM code.
In the default compilation settings the flag is commented out.
To include CP2K into compilation the user is required to enable the flag in the Makefile.
If the flag is omitted the PaPIM code will be compiled without CP2K and without the split communicator
parallelization scheme.
The latter is not used with any current analytic potential subroutine so it is omitted (for more
details on the potential subroutine see here).
We advise to compile the PaPIM code first successfully (by verifying the compilation by executing and checking
the standard tests) before recompiling and linking it to CP2K.
Upon adapting the Makefile, the code compilation is executed by command make in the ./source
sub-directory.
The executable PaPIM.exe is created at the same location upon successful compilation.
For module’s testing purposes the user is advised to have numdiff package installed before running the tests.
More details on the numdiff program package and its installation are available
here.
The PaPIM documentation is generated by executing the make command in the ./doc sub-directory.
Testing¶
Testing comprises of a set of tests for the analytic potentials and a set of tests for the CP2K interface
to PaPIM code.
Tests and all corresponding reference files are located in sub-directory ./tests.
All tests have to be performed in this sub-directory.
Details of the structure of the test files/sub-directories are explained in the PaPIM code testing
section.
CP2K tests and reference files are located in the sub-directory cp2k.
Tests are performed automatically in the ./tests sub-directory by executing the command:
./test.sh -c -n [number of cores]
The optional flag -c includes running of the CP2K tests and should only be used if PaPIM
code was compiled with CP2K.
If omitted only the tests using the analytic potential will be performed.
Flag -x omits the analytic potential tests and executes only the CP2K tests.
Flag -n [number of cores] controls the number of processor cores used in the tests.
Omitting this flag the tests will be performed on two processor cores (default value).
Because the number of trajectories for sampling in certain tests are limited to 20, the number
of processor cores should not exceed 20.
In all cases the CP2K potential is calculated on one processor core for each trajectory.
In order to change that, a different group_size variable should be specified manually in the
corresponding CONTROL files located in sub-directory of each test case.
Note that the total number of processor cores used in the tests should be divisible by the group_size
value (see details in Parallelization and Benchmarking section).
For comparison of generated output values with reference data the test script uses numdiff command
in order to compensate for small numerical differences.
By default the script looks first for the numdiff command on the system, and in case it fails to
locate it, the standard diff command will be used instead.
However, the user is warned that due to small numerical differences between generated output and corresponding
reference values the automated tests are most likely to fail.
A local numdiff package copy can be included in the test by specifying its absolute path.
For this and other options of the test script list them with the command ./test.sh -h.
Source Code¶
The full PaPIM code with the interface subroutine to the CP2K is located at: https://gitlab.e-cam2020.eu:10443/Quantum-Dynamics/PIM/tree/master/source.
Git is recommended for downloading the full copy of the code.
The main interface subroutines for linking PaPIM to CP2K are located in the Fortran module
file cp2k_module.f90. Corresponding commands used throughout the code can be located by searching
for the __USE_CP2K keyword.
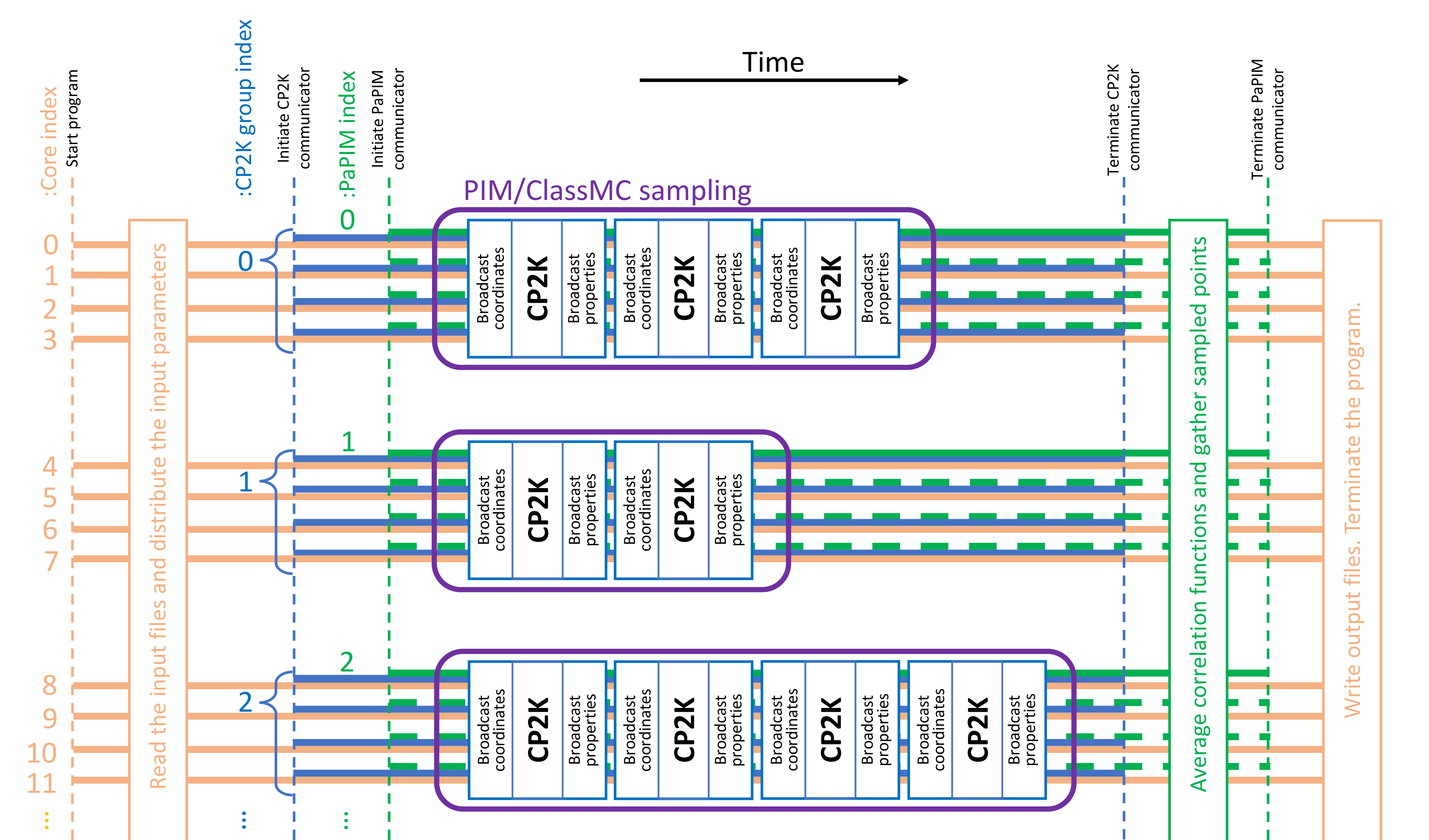
Parallelization scheme¶
Parallelization of linked PaPIM and CP2K codes is achieved with a MPI split communicator approach.
A separate communicator is given for the PaPIM code and for the CP2K part.
The latter is split into groups, each of a number of processor cores given by the group_size value.
Therefore, the number of trajectories which can be sampled simultaneously is given by the quotient of the
total number of used processor cores with the value of the group_size.
For the same reason the total number of cores must be divisible by the group_size value.
The figure below explains in a simplified graphical manner the parallelization used in the PaPIM code
linked to CP2K.
Source Code Documentation¶
The source code documentation is located in the ./doc sub-directory.
The documentation files (html and latex format) are generated by executing the make command in the ./doc
sub-directory.