n2p2 - Symmetry Function Memory Footprint Reduction¶
This module improves memory management in n2p2. More specifically, a new strategy to store symmetry function derivatives is implemented. In this way the memory footprint during training is drastically reduced.
Purpose of Module¶
Training high-dimensional neural network potentials (HDNNPs) means to minimize the error between predictions and the reference information in a data set of atomic configurations. There, the desired potential energy surface is supplied in the form of an energy per configuration and forces acting on each atom. Consider the HDNNP expression for forces

where 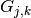 denotes the
denotes the 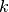 -th symmetry function of atom
-th symmetry function of atom
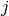 . Only the first expression
. Only the first expression 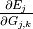 depends on the neural network weights and therefore changes during the
training process. The symmetry function derivatives with respect to atom
coordinates
depends on the neural network weights and therefore changes during the
training process. The symmetry function derivatives with respect to atom
coordinates 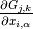 , however,
stay fixed for each atomic configuration in the data set. Given the high
computational cost of symmetry functions it is essential to pre-calculate and
store them in memory. While this strategy speeds up the training procedure
significantly [1] it also drastically increases the memory footprint, which
easily reaches more than 100 GB for common data set sizes.
, however,
stay fixed for each atomic configuration in the data set. Given the high
computational cost of symmetry functions it is essential to pre-calculate and
store them in memory. While this strategy speeds up the training procedure
significantly [1] it also drastically increases the memory footprint, which
easily reaches more than 100 GB for common data set sizes.
This module alters the core C++ library of n2p2 in order to reduce the memory
consumption of all depending applications and provides benchmark results
quantifying the improvement. The idea is to exploit that for specific
combinations of neighboring atoms 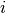 and
and 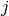 , the expression
, the expression
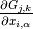 always equals zero.
Consider a three-component system with elements A, B and C. In addition, let
atoms
always equals zero.
Consider a three-component system with elements A, B and C. In addition, let
atoms 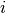 and
and 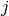 be of element A and B, respectively. Then, the
derivative of a symmetry function
be of element A and B, respectively. Then, the
derivative of a symmetry function 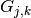 with signature B-C (i.e. only
sensitive to neighbor atoms of type C) with respect to
with signature B-C (i.e. only
sensitive to neighbor atoms of type C) with respect to 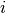 ’s coordinates
vanishes. Hence, by taking these element combination relations automatically into
account a significant portion of the memory usage can be avoided. Depending on
the symmetry function setup, savings of about 30 to 50% can be achieved for
typical systems. These improvements will be particularly helpful for developing
HDNNPs for coarse-grained models.
’s coordinates
vanishes. Hence, by taking these element combination relations automatically into
account a significant portion of the memory usage can be avoided. Depending on
the symmetry function setup, savings of about 30 to 50% can be achieved for
typical systems. These improvements will be particularly helpful for developing
HDNNPs for coarse-grained models.
Code changes cover most of the classes in the libnnp core library where they add functionality to identify relevant (nonzero) element combinations for the symmetry function derivative computation. Additional CI tests ensure that results are not affected.
Background Information¶
This module is based on n2p2, a C++ code for generation and application of neural network potentials used in molecular dynamics simulations. The source code and documentation are located here:
- n2p2 documentation: http://compphysvienna.github.io/n2p2/
- n2p2 source code: http://github.com/CompPhysVienna/n2p2
Building and Testing¶
Because the change in memory management affects the core library of n2p2
several applications shipped with n2p2 will benefit from reduced memory
consumption. However, the biggest effect can be observed during training with
the nnp-train application. In the src directory type
make nnp-train
to build this n2p2 tool (see the build documentation for more details).
Switch to one of the folders inside the examples/nnp-train directory and run
nnp-train (after a successful build the binary is copied to the bin
directory). The screen output will contain a section labelled SETUP: SYMMETRY
FUNCTION MEMORY which will highlight the memory savings.
The code changes from this module are already merged with the main repository of n2p2 (see pull request). The improved memory management is enabled by default when n2p2 is compiled. However, there are use cases (see this discussion) where the “full” memory layout is more desirable. Hence, a compilation flag allows to switch between the two choices. The documentation also shows benchmark results which demonstrate the potential memory savings.
Regression testing is implemented in n2p2 and automatically performed upon submission of a pull request via Travis CI. The log file showing the successful pass of all tests for the specific pull request can be found here.
Source Code¶
The easiest way to view the source code changes covered by this module is to use the GitHub pull request page. There, use the Files changed tab to review all changes.
| [1] | Singraber, A.; Morawietz, T.; Behler, J.; Dellago, C. Parallel Multistream Training of High-Dimensional Neural Network Potentials. J. Chem. Theory Comput. 2019, 15 (5), 3075–3092. |