E-CAM 2spaces_on_gpu module¶
The 2spaces_on_gpu module implements the 2-spaces algorithm on a GPU (see “Background Information” section). This algorithm is designed to move one-half of the polymer in one Monte-Carlo iteration. It also preserves the excluded volume constraints. An OpenCL implementation has been written so it can be used on CPUs or GPUs.
Purpose of Module¶
A polymer of size L is supposed to reach equilibrium
after a time of order 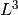 .
Therefore, it could be difficult to study the equilibrium
properties of large polymers.
The 2-spaces algorithm improves the efficiency
of each Monte-Carlo iteration by moving half of the polymer.
We can use the GPUs to take care of one of the sub-moves
in each Monte-Carlo step.
.
Therefore, it could be difficult to study the equilibrium
properties of large polymers.
The 2-spaces algorithm improves the efficiency
of each Monte-Carlo iteration by moving half of the polymer.
We can use the GPUs to take care of one of the sub-moves
in each Monte-Carlo step.
It is used in a scientific collaboration (ENS Lyon).
Background Information¶
Please consider reading the two research articles Massively Parallel Architectures and Polymer Simulation and Cellular automata for polymer simulation with application to polymer melts and polymer collapse including implications for protein folding for details about the method.
Building and Testing¶
I provide a simple Makefile as well as an
OpenCL
kernel and main source code to run the model.
You need C++11 in order to use pseudo-random number generator.
Before the compilation you can clean the previous build
with the make mrproper command.
Details about building, testing and running the code is available in the
2spaces_on_gpu GitLab repository.
Source Code¶
The source code and more information can be find on the 2spaces_on_gpu GitLab repository.