Particle Insertion Core¶
Purpose of the Module¶
This software module computes the change in free energy associated with the insertion or deletion of Lennard Jones particles in dilute or dense conditions in a variety of Thermodynamic Ensembles, where statistical sampling through molecular dynamics is performed under LAMMPS but will be extended to other molecular dynamics engines at a later date. Lennard-Jones type interactions are the key source of difficulty associated with particle insertion or deletion, which is why this module is a core module, as other interactions including Coulombic and bond, angle and dihedral interactions will be added in a second module. It differs from the main community approach used to date to compute such changes as it does not use soft-core potentials. Its key advantages over soft-core potentials are: (a) electrostatic interactions can in principle be performed simultaneously with particle insertion (this and other functionalities will be added in a new module); and, (b) essentially exact long-range dispersive interactions using dispersion Particle Mesh Ewald (PMME) or EWALD if desired can be selected at runtime by the user.
Background Information¶
Particle insertion can be used to compute the free energy associated with hydration/drying, the insertion of cavities in fluids/crystals, changes in salt levels, changes in solvent mixtures, and alchemical changes such as the mutation of amino-acids. in crystals. It can also be used to compute the free energy of solvent mixtures and the addition of salts, which is used in the purification processing industrially, for instance in the purification of pharmaceutical active ingredients. Particle insertion can in principle also be used to compute the free energy associated with changes in the pH, that is the proton transfer from a titratable site to the bulk, for example in water.
Our approach consists of rescaling the effective size of inserted atoms through a
parameter 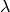 so that all interactions between
nserted atoms and interactions between inserted atoms and atoms already present in
the system are zero when
so that all interactions between
nserted atoms and interactions between inserted atoms and atoms already present in
the system are zero when 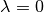 , creating at most an
integrable singularity which we can safely handle. In the context of Lennard-Jones
type pair potentials,
our approach at a mathematical level is similar to Simonson, who investigated the
mathematical conditions required to avoid the
singularity of insertion. It turns out
that a non-linear dependence of the
interaction on :math:’lambda’ between inserted
atoms and those already present is required (i.e. a simple linear dependence on
:math: ‘lambda’ necessarily introduces a singularity).
, creating at most an
integrable singularity which we can safely handle. In the context of Lennard-Jones
type pair potentials,
our approach at a mathematical level is similar to Simonson, who investigated the
mathematical conditions required to avoid the
singularity of insertion. It turns out
that a non-linear dependence of the
interaction on :math:’lambda’ between inserted
atoms and those already present is required (i.e. a simple linear dependence on
:math: ‘lambda’ necessarily introduces a singularity).
This module and upcoming modules include computing the free energy changes associated with the following applications
- hydration and drying;
- the addition of multiple molecules into a condenses environment;
- residue mutation and alchemy;
- constant pH simulations, this also will also exploit modules created in E-CAM work package 3 (quantum dynamics); and,
- free energy changes in chemical potentials associated with changes in solvent mixtures.
General Formulation¶
Consider a system consisting of 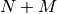 degrees of freedom and the Hamiltonian
degrees of freedom and the Hamiltonian
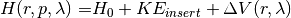
where 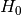 corresponds to an unperturbed Hamiltonian, and the perturbation
corresponds to an unperturbed Hamiltonian, and the perturbation
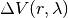 depends
nonlinearly on a control parameter
depends
nonlinearly on a control parameter 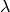 . The first set of N degrees of
freedom is denoted by A and the second
set of M degrees of freedom is denoted by B. To explore equilibrium properties of
the system, thermostats, and barostats
are used to sample either the NVT (canonical) ensemble or the NPT (Gibbs) ensemble. The
perturbation is devised so that
when
. The first set of N degrees of
freedom is denoted by A and the second
set of M degrees of freedom is denoted by B. To explore equilibrium properties of
the system, thermostats, and barostats
are used to sample either the NVT (canonical) ensemble or the NPT (Gibbs) ensemble. The
perturbation is devised so that
when 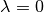 ,
, 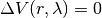 , B is in purely virtual.
When
, B is in purely virtual.
When 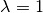 , B
corresponds to a fully physical augmentation of the original system.
, B
corresponds to a fully physical augmentation of the original system.
In the present software module, we consider only interaction Lennard Jones atoms.
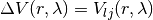
where for each inserted atom i
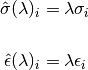
and the mixing rule for Van der Waals diameters and binding energy between different
atoms uses the geometric mean.
The dependence of 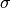 on
on 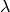 has the consequence that the mean
has the consequence that the mean
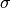 between a pair of inserted atoms scales as
between a pair of inserted atoms scales as 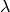 , but scales
as
, but scales
as 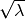 when one atom in the pair is
inserted and the other is already present. These choices of perturbations guarantees
that the particle insertion and deletion catastrophes are avoided.
when one atom in the pair is
inserted and the other is already present. These choices of perturbations guarantees
that the particle insertion and deletion catastrophes are avoided.
Algorithms¶
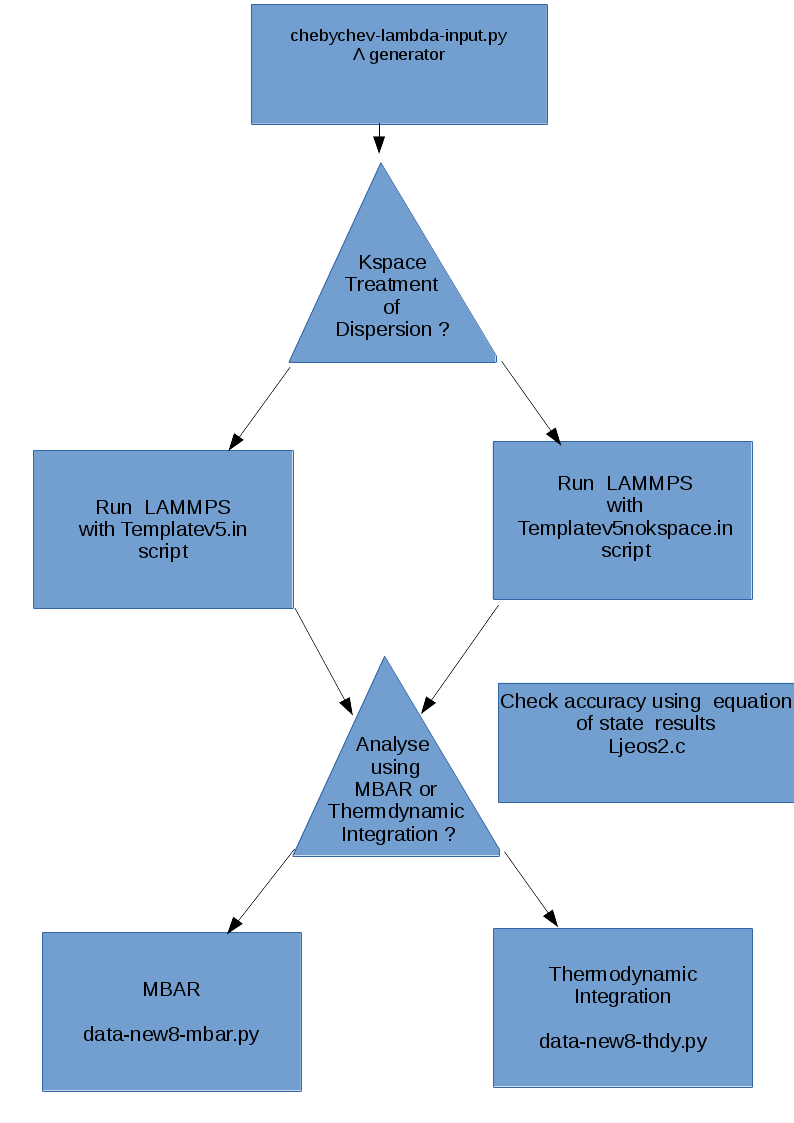
At the core of the PI core module there are four functions/codes. The first written in python generates the interpolation points which are the zero’s of suitably transformed Chebyshev functions.
The second code written ln LAMMPS scripting language performs the simulation in user-defined ensembles at the selected interpolation values of :math:’lambda’, at a user-specified frequency, computing two-point central difference estimates of derivatives of the potential energy needed for thermodynamic integration, computing the energy functions for all values of :math:’lambda’ in the context of MBAR. The user also specifies the locations of the inserted particles. The user also specifies whether Particle Mesh Ewald or EWALD should be used for dispersive interactions.
The third code written in python takes the output data from LAMMPS, prepares it so that free energy differences in the selected ensemble can be computed using MBAR provided by the pymbar suite of Python codes of the Chodera group.
The fourth code, also written in python take the LAMMPS output and performs the thermodynamic integration.
Source Code¶
All files can be found in the PIcore subdirectory of the
particle_insertion git repository.
Compilation and Linking¶
See PIcore README for full details.
Scaling and Performance¶
As the module uses LAMMPS, the performance and scaling of this module should essentially be the same, provided data for thermodynamic integration and MBAR are not generated too often, as is demonstated below. In the case of thermodynamic integration, this is due to the central difference approximation of derivatives, and in the case of MBAR, it is due to the fact that many virtual moves are made which can be extremely costly if the number of interpolating points is large. Also, when using PMME, the initial setup cost is computationally expensive, and should, therefore, be done as infrequently as possible. A future module in preparation will circumvent the use of central difference approximations of derivatives. The scaling performance of PI-CORE was tested on Jureca multi node. The results for weak scaling (where the number of core and the system size are doubled from 4 to 768 core) are as follows.
Weak Scaling:
| Number of MPI Core | timesteps/s |
|---|---|
| 4 | 1664.793 |
| 8 | 1534.013 |
| 16 | 1458.936 |
| 24 | 1454.075 |
| 48 | 1350.257 |
| 96 | 1301.325 |
| 192 | 1263.402 |
| 384 | 1212.539 |
| 768 | 1108.306 |
and for the strong scaling (where the number of core are doubled from 4 to 384 but the system size is fixed equal to 768 times the original system size considered for one core/processor for weak scaling) Strong Scaling:
| Number of MPI Core | timesteps/s |
|---|---|
| 4 | 9.197 |
| 8 | 17.447 |
| 16 | 34.641 |
| 24 | 53.345 |
| 48 | 104.504 |
| 96 | 204.434 |
| 192 | 369.178 |
| 384 | 634.022 |