Contact Concurrences¶
This module deals with the analysis of contacts between parts of biomolecules based on “contact concurrences,” i.e., what contacts occur simultaneously during a trajectory. This is useful when using contacts as a definition of a metastable state in a trajectory.
Purpose of Module¶
Contact frequencies, as developed in the module Contact Map, are a useful tool for studying biomolecular systems, such as binding/unbinding of a ligand from a protein. However, they suffer from one problem when trying to use them to define metastable states: since they are averaged over time, they don’t show time-dependent behavior. To identify a stable state, time-dependent behavior must be considered.
For example, a particular contact pair might have a frequency of 0.1 during a 100ns trajectory. But this could be achieved in several ways. If the contact events are randomly distributed through time, this contact probably isn’t characteristic of a metastable state. On the other hand, if the contact is constantly present during the last 10 ns (and not otherwise present), it might represent a metastable state. More importantly, there might be multiple contacts that are all present during those last 10 ns. Those concurrent contacts could be used to define a metastable state. This module helps identify and analyze those concurrent contacts by providing a tool to visualize them.
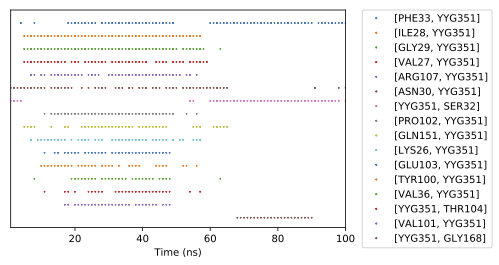
The figure shows the output of the contact concurrence visualization for the contacts between an inhibitor (labelled YYG) and various residues of the protein GSK3B. The plot shows when each contact occurred. The x-axis is time. Each dot represents that a specific contact pair is present at that time. The contact pairs are separated along the vertical axis.
This trajectory shows two groups of stable contacts between the protein and the ligand; i.e. there is a change in the stable state. This allows us to visually identify the contacts involved in each state. Both states involve the ligand being in contact with Phe33, but the earlier state includes contacts with Ile28, Gly29, etc., while the later state includes contacts with Ser32 and Gly168.
This is an important tool for identifying stable states based on long-lived groups of contacts, and is being used as part of the E-CAM pilot project on binding kinetics. It has also been used a part of a bachelor’s thesis project to develop an automated approach to identifying metastable intermediates during binding/unbinding processes.
Classes implemented in this module include:
Concurrence: Superclass for contact concurrence objects, enabling future custom concurrence types.AtomContactConcurrence: Contact concurrences for atom-atom contacts.ResidueContactConcurrence: Contact concurrences for residue-residue contacts (based on minimum distance between constituent atoms).ConcurrencePlotterandplot_concurrences: Class and convenience function (respectively) for making plots of contact concurrence.ContactsDict: Dict-like object giving access to atom or residue contacts based on string keys. Also addedContactObject.contactsproperty, which returns aContactsDictobject for theContactObject.
Background Information¶
This module is part of the contact_map project, which builds on tools from MDTraj.
Building and Testing¶
This module will be included in the 0.4 release of contact_map. After
that release, it can be easily installed with conda, using conda
install -c conda-forge contact_map, or conda install -c conda-forge
contact_map==0.4.0 for the first version that includes this module. To see
the current release, go to https://pypi.org/project/contact-map/#history.
Until the release, this module can only be installed through a developer
install of contact_map. This involves downloading the contact_map
repository, installing the requirements, and then installing the
contact_map package from source. Instructions can be found on the
installation page
of the contact_map documentation.
Once installed, tests are run using pytest. To check that the code has been
correctly installed, run python -c "import contact_map" from the command
line. To run the tests, install pytest and run the command py.test
--pyargs contact_map.
Source Code¶
The source code for this module is contained in the following pull requests
in the contact_map repository: