Maximum likelihood optimization of reaction coordinates¶
Authors: Clemens Moritz and Raffaela Cabriolu
This module implements an OpenPathSampling library that provides a maximum likelihood analysis to obtain an optimal reaction coordinate by combining multiple collective variables.
Purpose of Module¶
OpenPathSampling (OPS) is a software package that simulates complex processes using path sampling techniques and yields reactive trajectories between states of interest in a given system. However, such trajectories do not automatically lead to a physical understanding of the reaction mechanism. To gain such an understanding it is desirable to find a set of collective variables (CVs) that carry physically important information about the process.
The size and the shape of a crystalline cluster in a freezing liquid, the number of native contacts in a folding protein or bond length and bond angles in chemical reactions are examples of such CVs. The aim of this module is to find an optimized combination of multiple CVs into a single coordinate, that monitors the progress of the reaction. Such a coordinate is commonly called a reaction coordinate.
In methods used to study complex processes, having a good reaction coordinate either significantly improves their efficiency or it is a prerequisite for the reliability of their results.
The reaction coordinate is constructed by optimizing the likelihood function
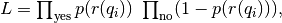
where r is a reaction coordinate model that combines several CVs, q_i, into a reaction coordinate and p is the probability model that maps this coordinate to a probability of having a successful outcome (yes). The definition of a successful outcome depends on the chosen probability model. Both r and p depend on a set of coefficients that are used to maximize L.
For more details on the method, please refer to the references given in Background Information.
Classes and objects implemented in this module:
TargetFunctionDescriptionclass. Wrapper around functions that carries additional information such as the number of parameters which shall be varied during subsequent optimization.REACTION_COORDINATE_MODELSdictionary of objects of the classTargetFunctionDescription. Collection of commonly used reaction coordinate models. At the moment two combinations of collective variables are available: a linear function, and a quadratic function. Additionally the user can define custom functions.PROBABILITY_MODELSdictionary of objects of the classTargetFunctionDescription. Collection of commonly used probability models. At the moment two functions are available: a sigmoidal function as a model for committor probabilities and a symmetric peaked function as a model for the probability of finding a transition path starting from the configuration r.MaxLikelihoodCVAnalysisclass. It implements the maximum likelihood analysis. There are two methods implemented: one for optimization based on committor probabilities, using anopenpathsampling.ShootingPointAnalysisobject (optimize_from_spa), and an other one where the user can perform optimizations for custom problems, using theoptimizemethod.
Background Information¶
This module is built on the OpenPathSampling library. More information about it are given in the following links:
- OPS documentation: http://openpathsampling.org
- OPS source code: http://github.com/openpathsampling/openpathsampling
Information about the method can be found in these publications:
- Peters, B. & Trout, B. L. “Obtaining reaction coordinates by likelihood maximization.” J. Chem. Phys. 125, 54108 (2006).
- Peters, B., Beckham, G. T. & Trout, B. L. “Extensions to the likelihood maximization approach for finding reaction coordinates.” J. Chem. Phys. 127, 34109 (2007).
- Peters, B. “Reaction Coordinates and Mechanistic Hypothesis Tests.” Annu. Rev. Phys. Chem. 67, annurev-physchem-040215-112215 (2016).
Testing¶
To test this module you need to download the source files package (see the Source Code section below) and install it using python setup.py install from the root directory of the package. In the ops_maxlikelihood/tests folder type nosetests to test the module using the nose package.
Examples¶
The example of the Maximum Likelihood module on the 2D toy model implemented in OPS is given in the directory examples. Open it using jupyter notebook ExampleMaximumLikelihood2DToyModel.ipynb (see Jupyter notebook documentation at http://jupyter.org/ for more details).
Source Code¶
Source code can be found at https://gitlab.e-cam2020.eu/Classical-MD_openpathsampling/MaxLikelihood