Interface Optimization¶
Author: Anastasiia Maslechko
This module consist of functions to evaluate the new values of the interface positions in Transition Interface Sampling (TIS) calculation. Two approaches are implemented (for more details please look in the Background Information).
Purpose of Module¶
In TIS calculations the full path space is investigated through several ensembles by putting the interfaces. For each interface during the simulation crossing probability is evaluated. At the end the reaction rate depends on the crossing probabilities obtained from each ensemble. Without a full knowledge of the reaction mechanism and information about free energy profile user most likely fails to set up manually initial setting, which are going to lead to the efficient usage of the software resources, and most importantly to low statistical errors in the simulation run.
One of the challenges in Transition Path Sampling is to define a proper collective variable (CV). Interface-optimization module is a way to improve code performance within selected CV.
Background Information¶
This module builds on OpenPathSampling, a Python package for path sampling simulations. To learn more about OpenPathSampling, you might be interested in reading:
- OPS documentation: http://openpathsampling.org
- OPS source code: http://github.com/openpathsampling/openpathsampling
In the Interface Optimization module a search of the optimal set of the interfaces is done in iterative manner. The key idea is described in the papers (please, see the references below), and the goal is to approach almost the same crossing probabilities in each ensemble:
- Ernesto E. Borrero and Fernando A. Escobedo. Optimizing the sampling and staging for simulations of rare events via forward flux sampling schemes. The Journal of Chemical Physics 129, 024115 (2008); doi: http://dx.doi.org/10.1063/1.2953325
- Ernesto E. Borrero, Marcus Weinwurm, and Christoph Dellago. Optimizing transition interface sampling simulations. The Journal of Chemical Physics 134, 244118 (2011); doi: http://dx.doi.org/10.1063/1.3601919
The first thing to do is to start the simulation for some number of cycles with initial guess (set of interfaces). On the basis of the obtained data analysis is done as follows: values of the current interface positions and the “crossing probabilities” (these are most likely non-converged values due to the least number of cycles) are used in the interface-optimization calculation. Note: termination of the simulation and getting of the interfaces and crossing probabilities must be done by an experimenter.
The next step is to build a mapping function f, which satisfies several properties:
- The form of the function helps to “equalize” the crossing probabilities on the next simulation run.
- It is bijective.
- It is monotonic in order to have inverse image.
The next step is to find the number of the interfaces (n_int) to satisfy users requirements. After that we need to divide a region from the starting point til 1 on the ordinate on the n_int - 1 intervals: by mapping of the obtained points on the function curve we can get a new set of interfaces.
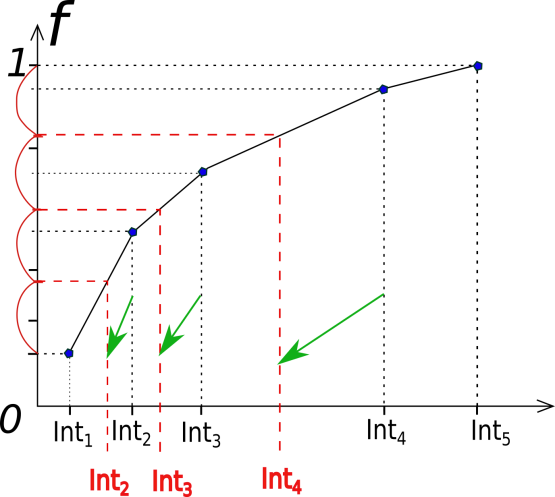
Important functions implemented in this module:
find_interface_and_cross: reads the output of the analyze-tool after a certain number of cycles (defined by the user). Object like TISTransition must be used as an input.save_n_interfaces: a method to calculate the new interface positions, using first approach: the number of interfaces kept the same, only the values of their positions are going to be changed.save_p_interfaces: a method to calculate the new interface positions, using second approach: the number of the interfaces might be changed, “expected” crossing probability for each ensemble has a predefined value.
Testing¶
Tests in OpenPathSampling use the nose package.
The tests for this module can be run by downloading its source code (check the section Source Code below), installing its requirements, and running the command nosetests in a root directory (or in particular for the file test_interface_optimization.py).
Examples¶
To check an example look for the file toy_mistis_interface_optimization.ipynb in the source code (or in the directory toy_model_mistis from the OPS-examples section). To open it use jupyter notebook toy_mistis_interface_optimization.ipynb (see Jupyter notebook documentation at http://jupyter.org/ for more details). In order to run it download the data from http://www.dropbox.com/s/qaeczkugwxkrdfy/toy_mistis_1k_OPS1.nc, as described inside of the example-file.
Source Code¶
The source code for this module can be found in: https://gitlab.e-cam2020.eu:10443/Classical-MD_openpathsampling/Interface.