Fine-graining: A Component of the Hierarchical Equilibration Strategy for Polymer Melts¶
The module is an implementation of the existing hierarchical strategy [1] for the equilibration of simple one-component polymer melts in ESPResSO++.
Purpose of Module¶
To study the properties of polymer melts by numerical simulations, equilibrated configurations must be prepared. However, the relaxation time for high molecular weight polymer melts is huge and increases, according to reptation theory, with the third power of the molecular weight. Hence, an effective method for decreasing the equilibration time is required. The hierarchical strategy pioneered in Ref. [1] is a particularly suitable way to do this. The present module provides a part of that method described below.
To decrease the relaxation time, microscopic monomers are coarse-grained (CG)
by mapping each subchain with 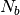 monomers onto a soft blob.
The CG system is then characterized by a much lower molecular weight and
thus is equilibrated quickly. One thus obtains a configuration that is
equilibrated on large scales but does not provide information about
the structure on smaller (i.e. more fine-grained (FG)) scales.
monomers onto a soft blob.
The CG system is then characterized by a much lower molecular weight and
thus is equilibrated quickly. One thus obtains a configuration that is
equilibrated on large scales but does not provide information about
the structure on smaller (i.e. more fine-grained (FG)) scales.
To obtain the latter, the resolution is step-by-step increased by recursively applying a fine-graining procedure to the previous (more coarse-grained) level. In such a fine-graining step, each CG polymer chain is replaced with a more fine-grained chain, by dividing a CG blob into several FG blobs.
The present module provides the python script which performs this fine-graining procedure. The implementation detail is in following below.
The microscopic configuration of
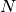 polymers consisted of
polymers consisted of 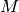 monomers is prepared. The system size
monomers is prepared. The system size 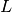 is determined by the number of density
is determined by the number of density 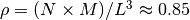 .
. 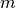 and
and 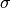 stands for the mass and the diameter of monomers.
stands for the mass and the diameter of monomers.We presuppose that equilibrated CG chain at
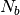 is already obtained.
is already obtained.The softblobs at
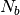 is divided into 2 softblobs at
is divided into 2 softblobs at 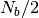 under the constraint conditions defined as
under the constraint conditions defined as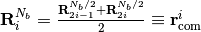 ,
,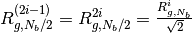 ,
,where
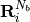 stands for
stands for
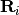 at
at 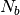 and
and 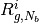 stand for
stand for 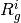 at
at 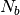 .
Namely, the center of mass of 2 softblobs at
.
Namely, the center of mass of 2 softblobs at 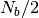 is identical
with the position of a softblob at
is identical
with the position of a softblob at 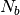 .
.For equilibrating a local configuration at
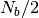 , NVT
MD simulation is performed.
, NVT
MD simulation is performed.In the beginning, a MD simulation takes into account bonding potential
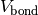 ,
,the potential for fluctuating radius of gyration
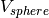 and the constrain potentials for center of mass described as
and the constrain potentials for center of mass described as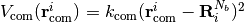 .
.Each
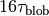 , MD simulation is including
the bending interactions
, MD simulation is including
the bending interactions 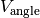
and non bonding interactions
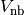 in this order.
in this order.Where
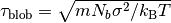 .
.After including all interactions, MD simulation is performed during
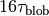 .
.In order to obtain the snapshot which has the ideal mean square internal distance (MSID)
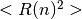 ,
MD simulation is continued to carry out. Where MSID
,
MD simulation is continued to carry out. Where MSID 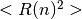 is defined as
is defined as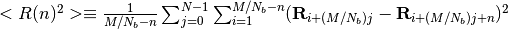 .
.This is calculated in each
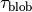 .
.After obtaining good snapshot at
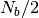 , fine-graining procedure is finished.
, fine-graining procedure is finished.
Background Information¶
The implementation of this module is based on ESPResSO++. You can learn about ESPResSO++ from the following links:
- ESPResSO++ documentation: http://espressopp.github.io/ESPResSo++.pdf
- ESPResSO++ source code: https://github.com/espressopp/espressopp
Building and Testing¶
Explanation of installation:
After installing this module, it can be tested according to the README file found under the following link:
Source Code¶
This module has been merged into ESPResSo++: