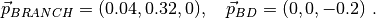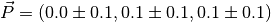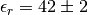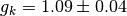Analysis of charge dipole moments in DL_MESO_DPD¶
Purpose of Module¶
This module, gen_dipole.f90, is a generalization of the dipole.f90 post-processing
utility of DL_MESO_DPD, the Dissipative Particle Dynamics (DPD) code from the DL_MESO package.
It processes trajectory (HISTORY) files to obtain the charge dipole moments
of all the (neutral) molecules in the system.
It produces files dipole_* containing the time evolution of relevant
quantities (see below). In the case of a single molecular species, it also prints
to the standard output the Kirkwood number 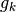 and the relative electric
permittivity
and the relative electric
permittivity 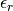 for this species, together with an estimate for
their errors (standard deviation).
for this species, together with an estimate for
their errors (standard deviation).
The module can be applied to systems including molecules with a generic charge structure, as long as each molecule is neutral (otherwise the charge dipole moment would be frame-dependent).
The charge dipole moment of a neutral molecule is 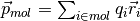 where
where 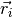 are the bead positions and
are the bead positions and 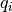 their charges. The
total charge dipole moment of the simulated volume
their charges. The
total charge dipole moment of the simulated volume 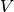 is
is
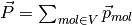 .
If more than one molecular species are present, one can split
.
If more than one molecular species are present, one can split 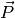 into the
different species’ contributions.
into the
different species’ contributions.
In general:
For any molecular species a file dipole_{molecule name} is produced, whose columns are
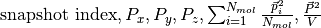 .
It is intended that for any quantity the contribution given from the
species {molecule name} is reported (i.e., the sums are restricted to
molecules of a single type).
.
It is intended that for any quantity the contribution given from the
species {molecule name} is reported (i.e., the sums are restricted to
molecules of a single type).
Possible uses of the output files are: monitoring the polarization in response to an external electric field, measuring the fluctuations in molecular/total charge dipole moments.
Extra output for a single molecular species:
The Kirkwood number for a pure system is
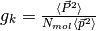 ,
where
,
where  indicates an average over trajectories.
If the dipoles’ orientations are not correlated, then
indicates an average over trajectories.
If the dipoles’ orientations are not correlated, then 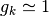 .
Also, the relative dielectric permittivity of the medium is calculated from linear response
theory:
.
Also, the relative dielectric permittivity of the medium is calculated from linear response
theory: 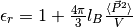 ,
where
,
where 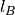 is Bjerrum length and tin-foil boundary conditions are used.
is Bjerrum length and tin-foil boundary conditions are used.
Background Information¶
The base code for this module is DL_MESO_DPD, the Dissipative Particle Dynamics code from the mesoscopic simulation package DL_MESO, developed by M. Seaton at Daresbury Laboratory. This open source code is available from STFC under both academic (free) and commercial (paid) licenses. The module is to be used with DL_MESO in its last released version, version 2.7 (dating December 2018).
A variant of this module for use with a previous version of DL_MESO,
version 2.6 (dating November 2015), can be found in the old-v2.6
directory [1].
Testing¶
The present module gen_dipole.f90 is compiled with the available Fortran90 compiler, e.g.:
gfortran -o gen_dipole.exe gen_dipole.f90
and the executable must be in the same directory of the HISTORY file to be analyzed. The user will be asked to provide the Bjerrum length used in single molecule simulations: all other information (including electric charges on all bead types) required for analyses is provided in the HISTORY file.
To input the Bjerrum length, one can either enter it from the keyboard or write it into a text file (say, input.txt) and run the program in this way:
gen_dipoleaf.exe < input.txt
We propose two tests to familiarize users with the utility and a third one on a physically relevant system.
The first two tests involve two (toy) molecular species: a branched one (four beads, T-shaped) and a simple dimer. All the beads carry charges. In the first case 10 molecules of each type are present and are followed for a few time steps. In the second case we suggest analyzing a single snapshot with just two molecules and all the beads sitting at user-defined positions (via a CONFIG file).
Four type of beads are used with charges 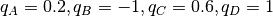 ;
the Bjerrum length is fixed as
;
the Bjerrum length is fixed as 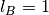 .
.
The bonding connections in the two molecules are pictorially given below:
B - A - C B - D
|
A
First test
Run the DL_MESO_DPD simulation on a single node (serial run) using the following CONTROL file:
Two kinds of molecules: branched and dimer
volume 3.0 3.0 3.0
temperature 1.0
cutoff 1.0
timestep 0.01
steps 1000
equilibration steps 0
traj 0 100 0
stats every 100
stack size 100
print every 100
job time 1000.0
close time 10.0
ensemble nvt mdvv
conf origin zero
ewald sum 1.0 5 5 5
bjerrum 1.0
smear gauss
smear length 0.5 equal
finish
and the FIELD file:
Two kinds of molecules: branched and dimer
SPECIES 4
A 1.0 0.2 0 0
B 1.0 -1.0 0 0
C 1.0 0.6 0 0
D 1.0 1.0 0 0
MOLECULES 2
BRANCH
nummols 10
beads 4
B 0.0 0.0 0.0
A 0.0 0.2 0.0
C 0.0 0.4 0.0
A 0.2 0.2 0.0
bonds 3
harm 1 2 5.0 0.25
harm 2 3 5.0 0.25
harm 2 4 5.0 0.25
finish
BD
nummols 10
beads 2
B 0.0 0.0 0.3
D 0.0 0.0 0.1
bonds 1
harm 1 2 5.0 0.25
finish
INTERACTIONS 4
A A dpd 25.0 1.0 4.5
B B dpd 25.0 1.0 4.5
C C dpd 25.0 1.0 4.5
D D dpd 25.0 1.0 4.5
CLOSE
Analyzing the HISTORY file with gen_dipole.exe, this output is printed on the standard output
nchist: 0 10 0 10
Number of snapshots: 11
<P_x>, <P_y>, <P_z>:
3.635198E-01 -9.224687E-02 5.177166E-01 8.410412E-01 3.127008E-01 5.555975E-01
error:
6.342381E-01 3.990649E-01 7.284979E-01 3.364702E-01 4.627111E-01 5.658327E-01
<P^2>/V:
4.857672E-01 2.793887E-01
error:
1.374485E-01 8.232245E-02
<p^2>:
1.381681E+00 8.502445E-01
error:
1.453820E-01 9.631156E-02
The first line shows the histogram of cluster sizes: in this case, it correctly gives 10 molecules of two beads, and 10 molecules of 4 beads. Since internally the module checks that each molecule is a connected cluster [2], this line should always give a histogram with the molecule sizes (up to the detected maximum number of beads per molecule).
The resulting dipole_BD file is
1 5.411139E-01 -3.599928E-01 -4.640084E-01 4.000000E-02 2.361863E-02
2 -2.554169E+00 -5.878279E-01 -1.010825E+00 1.310513E+00 2.922626E-01
3 1.110258E-01 1.629865E+00 5.048394E+00 9.721937E-01 1.042780E+00
4 -1.527498E+00 5.440944E-01 1.932210E+00 8.096611E-01 2.356565E-01
5 1.962445E+00 7.272408E-01 1.737201E+00 7.240524E-01 2.739976E-01
6 -2.179062E-01 -4.113665E-01 -1.396401E+00 8.849979E-01 8.024596E-02
7 1.552674E-01 2.753005E+00 -1.427860E-01 1.113470E+00 2.823531E-01
8 -8.580591E-01 1.490655E+00 7.832797E-01 1.145730E+00 1.322905E-01
9 2.232485E+00 2.690999E+00 -1.087863E+00 6.544034E-01 4.966263E-01
10 -1.225526E-01 3.129381E-01 1.879348E+00 7.360628E-01 1.349962E-01
11 -7.368671E-01 4.618429E-01 -1.166975E+00 9.616044E-01 7.844829E-02
and the dipole_BRANCH one is
1 -3.258640E-01 -4.896253E-01 -3.064526E-01 1.040000E-01 1.629013E-02
2 4.901658E+00 4.033448E+00 -1.381903E+00 1.525634E+00 1.563133E+00
3 -1.233831E+00 9.305151E-01 3.127786E+00 2.095572E+00 4.507868E-01
4 -3.059065E+00 -4.542448E-01 -1.278650E+00 1.559279E+00 4.147838E-01
5 1.576574E+00 -4.318085E+00 2.111019E+00 1.067546E+00 9.476980E-01
6 4.662262E-01 -1.342421E+00 2.268855E+00 1.601749E+00 2.654505E-01
7 -6.572569E-01 -9.276115E-02 5.649749E-01 1.480806E+00 2.814029E-02
8 -1.704146E+00 -1.273019E+00 7.975968E-01 1.078223E+00 1.911427E-01
9 9.061683E-01 1.749560E+00 -2.253253E-01 1.526127E+00 1.456620E-01
10 2.677888E-01 3.099173E+00 -6.472147E-01 1.494043E+00 3.739063E-01
11 2.860466E+00 3.852343E+00 -1.590978E+00 1.665513E+00 9.464453E-01
If instead the simulation is run on multiple nodes, only the results for the first snapshot will be unchanged (i.e., the first line of each dipole_* file). The other results will vary because different sequences of random numbers will be used by DL_MESO_DPD for the time evolution of the system.
Second test
Run DL_MESO_DPD using the same CONTROL and FIELD files as above, with the following changes:
- change “steps 1000” to “steps 1” (in CONTROL)
- change “nummols 10” to “nummols 1” (NB: appears twice in FIELD)
Also, use this CONFIG file that will initially align the molecule branches with the Cartesian axes:
Two kind of molecules, branched and dimer
0 1
3.0 0.0 0.0
0.0 3.0 0.0
0.0 0.0 3.0
B 1
0.0 0.0 0.0
A 2
0.0 0.2 0.0
C 3
0.0 0.4 0.0
A 4
0.2 0.2 0.0
B 5
0.0 0.0 0.3
D 6
0.0 0.0 0.1
where the identity of each bead is fixed by the FIELD file and is shown below:
B(1) - A(2) - C(3) B(5) - D(6)
|
A(4)
One can easily check that the dipole of each molecule is as expected (within machine precision):
The resulting dipole_BD file is
1 0.000000E+00 0.000000E+00 -2.000000E-01 4.000000E-02 1.481481E-03
and the dipole_BRANCH one is
1 4.000000E-02 3.200000E-01 2.220446E-16 1.040000E-01 3.851852E-03
The results of this test will not depend on the number of nodes used to run the simulation [3].
Third test: water in oil
Here we suggest considering a fluid made up of harmonically bonded dimers
 . Appropriately fixing the partial charges
. Appropriately fixing the partial charges 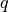 and the Bjerrum length
and the Bjerrum length 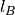 , this system
mimics water in an oil background as far as its dielectric properties
are concerned. For more details about this model, please see the page Test case: a dimer solvent.
, this system
mimics water in an oil background as far as its dielectric properties
are concerned. For more details about this model, please see the page Test case: a dimer solvent.
Run DL_MESO_DPD using the following CONTROL file:
DL_MESO charged harmonic dimers with dpd repulsion
volume 64.0
temperature 1.0
cutoff 1.0
timestep 0.01
steps 70000
equilibration steps 20000
traj 20000 100
stats every 100
stack size 100
print every 100
job time 7200.0
close time 100.0
ensemble nvt mdvv
ewald sum 1.0 5 5 5
bjerrum 42.0
smear gauss
smear length 0.5 equal
finish
and the FIELD file:
DL_MESO charged harmonic dimers with dpd repulsion
SPECIES 2
solp 1.0 0.46 0
solm 1.0 -0.46 0
MOLECULES 1
DIMER
nummols 96
beads 2
solp 0.0 0.0 0.0
solm 0.1 0.0 0.0
bonds 1
harm 1 2 5.0 0.0
finish
INTERACTIONS 3
solp solp dpd 25.0 1.0 4.5
solm solm dpd 25.0 1.0 4.5
solp solm dpd 25.0 1.0 4.5
CLOSE
Analyzing the HISTORY file with gen_dipole.exe, this output is printed to the standard output:
nchist: 0 96
Number of snapshots: 501
<P_x>, <P_y>, <P_z>:
-4.348820E-02 5.931873E-02 6.210429E-02
error:
1.035501E-01 9.685632E-02 9.780560E-02
<P^2>/V:
2.324029E-01
error:
8.812749E-03
<p^2>:
1.416901E-01
error:
4.351294E-04
kirkwood factor:
1.093480E+00
error:
4.482298E-02
Bjerrum length?
42.0
epsilon_r:
4.188645E+01
error:
1.550420E+00
In particular, we see that:
Please note that the error estimates are calculated assuming all the samples are
independent. From the results obtained in the testing case of the
module gen_dipoleaf.f90, one sees that the auto-correlation time of
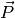 in this system is about 1-2 DPD time units, so the sampling choice
used here (trajectories are written every 100 time steps, i.e., at each DPD
time unit) seems reasonable, even if a little bit optimistic.
To confirm the reliability of the error estimate, one can carry out
another run with a different random number sequence (using the CONTROL file
directive seed) and see if the two results are compatible within error bars.
in this system is about 1-2 DPD time units, so the sampling choice
used here (trajectories are written every 100 time steps, i.e., at each DPD
time unit) seems reasonable, even if a little bit optimistic.
To confirm the reliability of the error estimate, one can carry out
another run with a different random number sequence (using the CONTROL file
directive seed) and see if the two results are compatible within error bars.
Source Code¶
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 179 180 181 182 183 184 185 186 187 188 189 190 191 192 193 194 195 196 197 198 199 200 201 202 203 204 205 206 207 208 209 210 211 212 213 214 215 216 217 218 219 220 221 222 223 224 225 226 227 228 229 230 231 232 233 234 235 236 237 238 239 240 241 242 243 244 245 246 247 248 249 250 251 252 253 254 255 256 257 258 259 260 261 262 263 264 265 266 267 268 269 270 271 272 273 274 275 276 277 278 279 280 281 282 283 284 285 286 287 288 289 290 291 292 293 294 295 296 297 298 299 300 301 302 303 304 305 306 307 308 309 310 311 312 313 314 315 316 317 318 319 320 321 322 323 324 325 326 327 328 329 330 331 332 333 334 335 336 337 338 339 340 341 342 343 344 345 346 347 348 349 350 351 352 353 354 355 356 357 358 359 360 361 362 363 364 365 366 367 368 369 370 371 372 373 374 375 376 377 378 379 380 381 382 383 384 385 386 387 388 389 390 391 392 393 394 395 396 397 398 399 400 401 402 403 404 405 406 407 408 409 410 411 412 413 414 415 416 417 418 419 420 421 422 423 424 425 426 427 428 429 430 431 432 433 434 435 436 437 438 439 440 441 442 443 444 445 446 447 448 449 450 451 452 453 454 455 456 457 458 459 460 461 462 463 464 465 466 467 468 469 470 471 472 473 474 475 476 477 478 479 480 481 482 483 484 485 486 487 488 489 490 491 492 493 494 495 496 497 498 499 500 501 502 503 504 505 506 507 508 509 510 511 512 513 514 515 516 517 518 519 520 521 522 523 524 525 526 527 528 529 530 531 532 533 534 535 536 537 538 539 540 541 542 543 544 545 546 547 548 549 550 551 552 553 554 555 556 557 558 559 560 561 562 563 564 565 566 567 568 569 570 571 572 573 574 575 576 577 578 579 580 581 582 583 584 585 586 587 588 589 590 591 592 593 594 595 596 597 598 599 600 601 602 603 604 605 606 607 608 609 610 611 612 613 614 615 616 617 618 619 620 621 622 623 624 625 626 627 628 629 630 631 632 633 634 635 636 637 638 639 640 641 642 643 644 645 646 647 648 649 650 651 652 653 654 655 656 657 658 659 660 661 662 663 664 665 666 667 668 669 670 671 672 673 674 675 676 677 678 679 680 681 682 683 684 685 686 687 688 689 690 691 692 693 694 695 696 697 698 699 700 701 702 703 704 705 706 707 708 709 710 | PROGRAM gen_dipole
!***********************************************************************************
! module to analyze charge dipole moments in DL_MESO
!
! authors: m. a. seaton and s. chiacchiera, February 2017 (amended January 2021)
!**********************************************************************************
IMPLICIT none
INTEGER, PARAMETER :: dp = SELECTED_REAL_KIND (15, 307)
INTEGER, PARAMETER :: li = SELECTED_INT_KIND (12)
INTEGER, PARAMETER :: ntraj=10
INTEGER, PARAMETER :: endversion = 1
REAL(KIND=dp), PARAMETER :: pi=3.141592653589793_dp
CHARACTER(80) :: text
CHARACTER(8), ALLOCATABLE :: namspe (:), nammol (:)
INTEGER, ALLOCATABLE :: ltp (:), ltm (:), mole (:), bndtbl (:,:)
INTEGER, ALLOCATABLE :: nbdmol (:), readint (:)
INTEGER, ALLOCATABLE :: visit (:), from (:)
INTEGER :: nrtout
INTEGER :: chain, imol, ioerror, i, numtraj, j, k, nmoldef, ibond, nbdmolmx
INTEGER :: nspe, nbeads, nusyst, nmbeads, nsyst, numbond, global, species, molecule
INTEGER :: nummol, lfrzn, rnmol, keytrj, srfx, srfy, srfz
INTEGER :: nav
INTEGER :: endver, Dlen, nstep, framesize, lend, leni
INTEGER(KIND=li) :: filesize, currentpos, lend_li, leni_li
REAL(KIND=dp), ALLOCATABLE :: xxx (:), yyy (:), zzz (:), readdata (:)
REAL(KIND=dp), ALLOCATABLE :: nmol (:), chg (:), molchg (:)
REAL(KIND=dp), ALLOCATABLE :: dipx_box (:), dipy_box (:), dipz_box (:)
REAL(KIND=dp), ALLOCATABLE :: dip2_box (:), dip2_ave (:)
REAL(KIND=dp), ALLOCATABLE :: dip2_err (:)
REAL(KIND=dp), ALLOCATABLE :: sum_dipx_box (:), sum_dipy_box (:), sum_dipz_box (:)
REAL(KIND=dp), ALLOCATABLE :: sum_dipx_box2 (:), sum_dipy_box2 (:), sum_dipz_box2 (:)
REAL(KIND=dp), ALLOCATABLE :: sum_dip2_box (:), sum_dip_box4 (:)
REAL(KIND=dp), ALLOCATABLE :: dipx_box_ave (:), dipy_box_ave (:), dipz_box_ave (:)
REAL(KIND=dp), ALLOCATABLE :: dipx_box2_ave (:), dipy_box2_ave (:), dipz_box2_ave (:)
REAL(KIND=dp), ALLOCATABLE :: dipx_box_err (:), dipy_box_err (:), dipz_box_err (:)
REAL(KIND=dp), ALLOCATABLE :: dip_box2_ave (:), dip_box2_err (:), dip_box4_ave (:)
REAL(KIND=dp), ALLOCATABLE :: gk (:), gk_err (:)
REAL(KIND=dp), ALLOCATABLE :: sum_dip2_box2 (:)
REAL(KIND=dp), ALLOCATABLE :: epsilon_r (:), epsilon_r_err(:)
REAL(KIND=dp) :: bjerelec
REAL(KIND=dp) :: volm, dimx, dimy, dimz, shrdx, shrdy, shrdz
REAL(KIND=dp) :: amass, rcii
REAL(KIND=dp) :: time
LOGICAL :: eof, swapend, bigend
! determine number of bytes for selected double precision and integer kinds
! (the default SELECTED_REAL_KIND (15, 307) should return 8 bytes)
lend = STORAGE_SIZE (1.0_dp) / 8
leni = BIT_SIZE (1) / 8
lend_li = INT (lend, KIND=li)
leni_li = INT (leni, KIND=li)
! check endianness of machine
bigend = (IACHAR(TRANSFER(1,"a"))==0)
! determine if HISTORY file exists, which endianness to use,
! if type of real is correct
INQUIRE (file = 'HISTORY', EXIST = eof)
IF (.NOT. eof) THEN
PRINT *, "ERROR: cannot find HISTORY file"
STOP
END IF
OPEN (ntraj, file = 'HISTORY', access = 'stream', form = 'unformatted', status = 'unknown')
swapend = .false.
READ (ntraj) endver, Dlen
IF (endver/=endversion) THEN
swapend = .true.
CLOSE (ntraj)
IF (bigend) THEN
OPEN (ntraj, file = 'HISTORY', access = 'stream', form = 'unformatted', status = 'unknown', convert = 'little_endian')
ELSE
OPEN (ntraj, file = 'HISTORY', access = 'stream', form = 'unformatted', status = 'unknown', convert = 'big_endian')
END IF
READ (ntraj) endver, Dlen
IF (endver/=endversion) THEN
PRINT *, "ERROR: corrupted HISTORY file or created with incorrect version of DL_MESO"
STOP
END IF
END IF
IF (Dlen/=lend) THEN
PRINT *, "ERROR: incorrect type of real number used in HISTORY file"
PRINT *, " recompile gen_dipole.f90 with reals of ", Dlen, " bytes"
STOP
END IF
! read file size, number of frames and timestep numbers
READ (ntraj) filesize, numtraj, nstep
! Read where the number of beads, molecules and bonds are determined
! Arrays are filled with names of particles and molecules
READ (ntraj) text
READ (ntraj) nspe, nmoldef, nusyst, nsyst, numbond, keytrj, srfx, srfy, srfz
IF (numbond==0) THEN
PRINT *, 'ERROR: no molecules in trajectory data!'
STOP
END IF
IF (srfx==1 .OR. srfy==1 .OR. srfz==1) THEN
WRITE (*,*) "ERROR: Systems under shear not yet implemented!"
STOP
END IF
framesize = (keytrj+1) * 3
ALLOCATE (readint (1:nsyst), readdata (1:framesize))
! get number of beads to be tracked when reading trajectory file (molecular beads)
nmbeads = nsyst - nusyst
ALLOCATE (namspe (nspe), nammol (nmoldef))
ALLOCATE (xxx (1:nmbeads), yyy (1:nmbeads), zzz (1:nmbeads))
ALLOCATE (ltp (1:nmbeads), ltm (1:nmbeads), mole (1:nmbeads))
ALLOCATE (nmol (1:nmoldef), nbdmol (1:nmoldef))
ALLOCATE (chg (nspe))
ALLOCATE (bndtbl (numbond, 2))
ALLOCATE (visit (nmbeads), from (nmbeads))
DO i = 1, nspe
READ (ntraj) namspe (i), amass, rcii, chg (i), lfrzn
END DO
DO i = 1, nmoldef
READ (ntraj) nammol (i)
END DO
! reading of bead species and molecule types
nummol = 0 !counter for number of molecules
ibond = 0 !counter for bonds
DO i = 1, nsyst
READ (ntraj) global, species, molecule, chain
IF (global>nusyst .AND. global<=nsyst) THEN
ltp (global-nusyst) = species
ltm (global-nusyst) = molecule
mole (global-nusyst) = chain
nummol = MAX (nummol, chain)
END IF
END DO
! reading of bond tables
IF (numbond>0) THEN
DO i = 1, numbond
READ (ntraj) bndtbl (i, 1), bndtbl (i, 2)
END DO
END IF
bndtbl = bndtbl - nusyst
! reached end of header: find current position in file
INQUIRE (unit=ntraj, POS=currentpos)
! determine numbers of molecules and beads per molecule type
nmol = 0.0_dp
nbdmol = 0
chain = 0
imol = 0 ! necessary to avoid out of bounds
DO i = 1, nmbeads
IF (mole (i) /= chain) THEN
chain = mole (i)
imol = ltm (i)
nmol (imol) = nmol (imol) + 1.0_dp
END IF
IF (imol > 0) nbdmol (imol) = nbdmol (imol) + 1
END DO
DO i = 1, nmoldef
rnmol = NINT (nmol (i))
IF (rnmol>0) THEN
nbdmol (i) = nbdmol (i) / rnmol
END IF
END DO
nbdmolmx = MAXVAL (nbdmol (1:nmoldef))
! obtain connectivity information (needed only once)
CALL connect (nmbeads, numbond, bndtbl, nbdmolmx, visit, from)
! Checking for charge neutrality of all molecules
ALLOCATE (molchg (nummol))
molchg (:) = 0._dp
DO i = 1, nmbeads
imol = mole (i)
molchg (imol) = molchg (imol) + chg (ltp (i))
END DO
DO i = 1, nummol
IF (ABS (molchg (i)) > 1.d-16) THEN
WRITE (*,*) "molecule number",i," is not neutral! (The dipole moment is frame-dependent)"
WRITE (*,*) "its charge is=", molchg (i)
WRITE (*,*) "its type is=", nammol (i)
STOP
ENDIF
END DO
CALL check_molecules !checks that beads are labelled are as expected
!reading trajectories and computing charge dipole moments
ALLOCATE (dipx_box (nmoldef), dipy_box (nmoldef), dipz_box (nmoldef))
ALLOCATE (dip2_box (nmoldef))
ALLOCATE (sum_dipx_box (nmoldef), sum_dipy_box (nmoldef), sum_dipz_box (nmoldef))
ALLOCATE (sum_dipx_box2 (nmoldef), sum_dipy_box2 (nmoldef), sum_dipz_box2 (nmoldef))
ALLOCATE (sum_dip2_box (nmoldef), sum_dip_box4 (nmoldef))
ALLOCATE (dipx_box_ave (nmoldef), dipy_box_ave (nmoldef), dipz_box_ave (nmoldef))
ALLOCATE (dip2_ave (nmoldef), dip2_err (nmoldef))
ALLOCATE (dipx_box2_ave (nmoldef), dipy_box2_ave (nmoldef), dipz_box2_ave (nmoldef))
ALLOCATE (dipx_box_err (nmoldef), dipy_box_err (nmoldef), dipz_box_err (nmoldef))
ALLOCATE (dip_box2_ave (nmoldef), dip_box2_err (nmoldef), dip_box4_ave (nmoldef))
ALLOCATE (sum_dip2_box2 (nmoldef))
ALLOCATE (gk (nmoldef), gk_err (nmoldef))
ALLOCATE (epsilon_r (nmoldef), epsilon_r_err(nmoldef))
! Open and write output file(s)
nrtout = ntraj + 1
DO j = 1, nmoldef
OPEN (nrtout+j-1, file = 'dipole_'//nammol(j), status ='replace')
END DO
eof = .false.
nav = 0
sum_dipx_box = 0.0_dp
sum_dipy_box = 0.0_dp
sum_dipz_box = 0.0_dp
sum_dip2_box = 0.0_dp
sum_dip_box4 = 0.0_dp
sum_dip2_box2 = 0.0_dp
sum_dipx_box2 = 0.0_dp
sum_dipy_box2 = 0.0_dp
sum_dipz_box2 = 0.0_dp
DO k = 1, numtraj
READ (ntraj, IOSTAT=ioerror) time, nbeads, dimx, dimy, dimz, shrdx, shrdy, shrdz
IF (ioerror/=0) THEN
eof = .true.
IF (k==1) THEN
WRITE (*,*) 'ERROR: cannot find trajectory data in HISTORY files'
STOP
END IF
EXIT
END IF
nav = nav + 1
volm = dimx * dimy * dimz
READ (ntraj) readint (1:nsyst)
DO i = 1, nsyst
global = readint (i)
READ (ntraj) readdata (1:framesize)
IF (global>nusyst .AND. global<=nsyst) THEN
xxx (global-nusyst) = readdata (1)
yyy (global-nusyst) = readdata (2)
zzz (global-nusyst) = readdata (3)
END IF
END DO
CALL compute_charge_dipoles (dipx_box, dipy_box, dipz_box, dip2_box)
DO j = 1, nmoldef
WRITE (nrtout+j-1, '(1p,I8,5(2x,e14.6))') k, dipx_box(j), dipy_box(j), dipz_box(j), dip2_box(j) / nmol(j) , &
(dipx_box(j)**2 + dipy_box(j)**2 + dipz_box(j)**2) / volm
END DO
sum_dipx_box = sum_dipx_box + dipx_box
sum_dipy_box = sum_dipy_box + dipy_box
sum_dipz_box = sum_dipz_box + dipz_box
sum_dipx_box2 = sum_dipx_box2 + dipx_box * dipx_box
sum_dipy_box2 = sum_dipy_box2 + dipy_box * dipy_box
sum_dipz_box2 = sum_dipz_box2 + dipz_box * dipz_box
sum_dip_box4 = sum_dip_box4 + (dipx_box**2 + dipy_box**2 + dipz_box**2)**2
sum_dip2_box = sum_dip2_box + dip2_box
sum_dip2_box2 = sum_dip2_box2 + dip2_box * dip2_box
END DO ! end of loop over trajectories
dipx_box_ave = sum_dipx_box/REAL(nav, KIND=dp)
dipy_box_ave = sum_dipy_box/REAL(nav, KIND=dp)
dipz_box_ave = sum_dipz_box/REAL(nav, KIND=dp)
dipx_box2_ave = sum_dipx_box2/REAL(nav, KIND=dp)
dipy_box2_ave = sum_dipy_box2/REAL(nav, KIND=dp)
dipz_box2_ave = sum_dipz_box2/REAL(nav, KIND=dp)
dip2_ave = sum_dip2_box(:)/REAL(nav, KIND=dp)/REAL(nmol(:), KIND=dp)
dip_box2_ave = dipx_box2_ave + dipy_box2_ave + dipz_box2_ave
dip_box4_ave = sum_dip_box4/REAL(nav, KIND=dp)
dipx_box_err = SQRT((dipx_box2_ave - dipx_box_ave**2)/REAL(nav, KIND=dp))
dipy_box_err = SQRT((dipy_box2_ave - dipy_box_ave**2)/REAL(nav, KIND=dp))
dipz_box_err = SQRT((dipz_box2_ave - dipz_box_ave**2)/REAL(nav, KIND=dp))
dip_box2_err = sqrt((dip_box4_ave - dip_box2_ave**2)/REAL(nav, KIND=dp))
! Error on dip2_ave is computed considering each snapshot as a sample
dip2_err = sum_dip2_box2 / dble(nav * nmol ** 2) - dip2_ave ** 2
dip2_err = sqrt (dip2_err / REAL(nav, KIND=dp))
WRITE (*,*) "Number of snapshots: ",nav
WRITE (*,*) "<P_x>, <P_y>, <P_z>:"
WRITE (*,98) dipx_box_ave, dipy_box_ave, dipz_box_ave
WRITE (*,*) "error:"
WRITE (*,98) dipx_box_err, dipy_box_err, dipz_box_err
WRITE (*,*) "<P^2>/V:"
WRITE (*,98) dip_box2_ave/volm
WRITE (*,*) "error:"
WRITE (*,98) dip_box2_err/volm
WRITE (*,*) "<p^2>:"
WRITE (*,98) dip2_ave
WRITE (*,*) "error:"
WRITE (*,98) dip2_err
IF (nmoldef == 1) THEN
gk = dip_box2_ave / dip2_ave / REAL(nmol, KIND=dp)
gk_err = (dip_box2_err / dip_box2_ave + dip2_err / dip2_ave) * gk
WRITE (*,*) "kirkwood factor:"
WRITE (*,98) gk
WRITE (*,*) "error:"
WRITE (*,98) gk_err
WRITE (*,*) "Bjerrum length?"
READ (*,*) bjerelec
epsilon_r = 1.0_dp + 4.0_dp * pi / 3.0_dp * bjerelec * dip_box2_ave / volm
epsilon_r_err = 4.0_dp * pi / 3.0_dp * bjerelec * dip_box2_err / volm
WRITE (*,*) "epsilon_r:"
WRITE (*,98) epsilon_r
WRITE (*,*) "error:"
WRITE (*,98) epsilon_r_err
ENDIF
! Close the trajectory file
CLOSE (ntraj)
!close output files
DO j = 1, nmoldef
CLOSE (nrtout+j-1)
END DO
DEALLOCATE (readint, readdata)
DEALLOCATE (namspe, nammol)
DEALLOCATE (xxx, yyy, zzz)
DEALLOCATE (ltp, ltm, mole)
DEALLOCATE (nmol, nbdmol)
DEALLOCATE (chg, molchg)
DEALLOCATE (dipx_box, dipy_box, dipz_box)
DEALLOCATE (dip2_box)
DEALLOCATE (sum_dipx_box, sum_dipy_box, sum_dipz_box)
DEALLOCATE (sum_dipx_box2 , sum_dipy_box2 , sum_dipz_box2)
DEALLOCATE (sum_dip2_box, sum_dip_box4, sum_dip2_box2)
DEALLOCATE (dipx_box_ave , dipy_box_ave , dipz_box_ave)
DEALLOCATE (dip2_ave, dip2_err)
DEALLOCATE (dipx_box2_ave , dipy_box2_ave , dipz_box2_ave)
DEALLOCATE (dipx_box_err , dipy_box_err , dipz_box_err)
DEALLOCATE (dip_box2_ave, dip_box2_err, dip_box4_ave)
DEALLOCATE (gk, gk_err)
DEALLOCATE (epsilon_r, epsilon_r_err)
DEALLOCATE (bndtbl)
DEALLOCATE (visit, from)
98 FORMAT(1p,9(e13.6,3x))
CONTAINS
SUBROUTINE check_molecules
!*************************************************************************************
! subroutine to check molecular content and labelling
!
! authors: s. chiacchiera, February 2017
!*************************************************************************************
IMPLICIT NONE
INTEGER i, j, k, tm, tp, imol, im, ibd
INTEGER mxmolsize
INTEGER, ALLOCATABLE :: molbeads (:,:)
mxmolsize = 0
DO i = 1, nmoldef
mxmolsize = MAX (nbdmol(i), mxmolsize)
END DO
ALLOCATE (molbeads (nmoldef, mxmolsize))
molbeads (:,:) = 0
imol = 0
ibd = 0
DO i = 1, nmoldef
DO j = 1, NINT (nmol(i))
imol = imol +1
DO k = 1, nbdmol(i)
ibd = ibd +1
tm = ltm (ibd)
tp = ltp (ibd)
im = mole (ibd)
IF (j==1) THEN
molbeads (i, k) = tp
ELSE
IF (molbeads (i, k) /= tp) THEN
WRITE (*,*) "ERROR: Problem with molecular content!"
STOP
ENDIF
ENDIF
IF (tm/=i.OR.im/=imol)THEN
WRITE (*,*) "ERROR: Problem with molecules labels!"
STOP
ENDIF
END DO
END DO
END DO
IF (imol/=nummol) THEN
WRITE (*,*) "ERROR: imol and nummol differ!"
STOP
ENDIF
DEALLOCATE (molbeads)
RETURN
END SUBROUTINE check_molecules
SUBROUTINE compute_charge_dipoles (dipx_box, dipy_box, dipz_box, dip2_box)
!*************************************************************************************
! subroutine to compute charge dipole moments
!
! authors: m. a. seaton and s. chiacchiera, February 2017
!
! input: xxx, yyy, zzz (at a given time step) and chg
! input: visit and from (obtained using connect)
! output: the x,y,z components of the total dipole, sum p_i^2/N_mol for each molecule
! type (at a given time step)
!*************************************************************************************
IMPLICIT NONE
INTEGER i, j, k, tm, tp, imol, ibd, count, ipr
REAL(KIND=dp), DIMENSION(nmoldef) :: dipx_box, dipy_box, dipz_box
REAL(KIND=dp), DIMENSION(nmoldef) :: dip2_box
REAL(KIND=dp) :: x, y, z, dx, dy, dz, xpre, ypre, zpre
REAL(KIND=dp) :: dipx, dipy, dipz, dip2
REAL(KIND=dp), DIMENSION(nmbeads) :: xabs, yabs, zabs
dipx_box (:) = 0._dp
dipy_box (:) = 0._dp
dipz_box (:) = 0._dp
dip2_box (:) = 0._dp
imol = 0
count = 0
! xabs = 0._dp ! just to keep it clean
! yabs = 0._dp
! zabs = 0._dp
DO i = 1, nmoldef
tm = i
DO j = 1, NINT (nmol(i))
imol = imol + 1
dipx = 0._dp ! dipole of a SINGLE molecule
dipy = 0._dp
dipz = 0._dp
DO k = 1, nbdmol(i)
count = count + 1
ibd = visit (count)
ipr = from (count)
IF (ipr /= 0) THEN
xpre = xabs (ipr)
ypre = yabs (ipr)
zpre = zabs (ipr)
ELSE
IF (k == 1) THEN
xpre = 0._dp
ypre = 0._dp
zpre = 0._dp
ELSE
WRITE (*,*) "Unconnected molecule!"
STOP
ENDIF
ENDIF
tp = ltp (ibd)
dx = xxx (ibd) - xpre
dy = yyy (ibd) - ypre
dz = zzz (ibd) - zpre
dx = dx - dimx * ANINT (dx/dimx)
dy = dy - dimy * ANINT (dy/dimy)
dz = dz - dimz * ANINT (dz/dimz)
x = xpre + dx
y = ypre + dy
z = zpre + dz
dipx = dipx + x * chg (tp)
dipy = dipy + y * chg (tp)
dipz = dipz + z * chg (tp)
xabs (ibd) = x
yabs (ibd) = y
zabs (ibd) = z
END DO
dipx_box (tm) = dipx_box (tm) + dipx
dipy_box (tm) = dipy_box (tm) + dipy
dipz_box (tm) = dipz_box (tm) + dipz
dip2 = dipx * dipx + dipy * dipy + dipz * dipz
dip2_box (tm) = dip2_box (tm) + dip2
END DO
END DO
IF (imol/=nummol) THEN
WRITE (*,*) "ERROR: imol and nummol differ!"
STOP
ENDIF
RETURN
END SUBROUTINE compute_charge_dipoles
End PROGRAM gen_dipole
SUBROUTINE connect (nbeads, nbonds, bndtbl, mxmolsize, visit, from)
!**********************************************************************
! Analyzes all the bonds (bndtbl) to obtain a schedule (visit, from)
! to visit the beads so that each cluster is visited along a connected
! path. "visit" gives the order to include beads, "from" gives the bead
! to attach them to.
! (Note: vocabulary from infection propagation used to move along
! clusters)
!
! author: s. chiacchiera, February 2017
! amended: m. a. seaton, January 2021
!**********************************************************************
IMPLICIT none
INTEGER, INTENT (INOUT) :: bndtbl (nbonds,2)
INTEGER, INTENT (IN) :: nbeads, nbonds
INTEGER, INTENT (IN) :: mxmolsize
INTEGER :: ic, i, k, nn, nclu, nper, lab, ref, count
INTEGER, ALLOCATABLE :: firstnn (:), lastnn (:), deg (:)
INTEGER, ALLOCATABLE :: labnn (:)
INTEGER, ALLOCATABLE :: state (:)
INTEGER, ALLOCATABLE :: perlab (:), perref (:)
INTEGER, ALLOCATABLE :: nchist (:)
INTEGER, INTENT (OUT) :: visit (nbeads), from (nbeads)
ALLOCATE (firstnn (nbeads), lastnn (nbeads), deg (nbeads), labnn (2*nbonds))
ALLOCATE (state (nbeads))
ALLOCATE (perlab (nbeads), perref (nbeads))
ALLOCATE (nchist (mxmolsize))
!-----------------------------------------------------------------------
CALL organize (nbeads, nbonds, labnn, firstnn, lastnn, deg)
!-----------------------------------------------------------------------
state (:) = 0
nchist (:) = 0
visit (:) = 0
from (:) = 0
count = 0
!-----------------------------------------------------------------------
ic = 0
!-----------------------------------------------------------------------
DO WHILE (ic < nbeads) ! ic = label of bead used to "grow" a cluster
ic = ic + 1
IF( state (ic) /= 0) THEN
WRITE (*,*) "ERROR: labels are not as expected!"
STOP
END IF
nclu = 1
count = count + 1
visit (ic) = ic
IF (deg (ic) == 0) THEN
state (ic) = -1
IF (nclu <= mxmolsize) nchist (nclu) = nchist (nclu) +1
CYCLE
END IF
state (ic) = 1 ! ic is "infected"
! nearest neighbours of ic are marked as "goint to be infected" -> a.k.a. perimeter
nper = 0
perlab (:) = 0
perref (:) = 0
DO k = firstnn (ic), lastnn (ic)
nn = labnn (k)
IF( state (nn) /= 0) THEN
WRITE (*,*) "ERROR: labels are not as expected!"
STOP
END IF
nper = nper + 1
perlab (nper) = nn !new bead in perimeter
perref (nper) = ic !its reference bead (origin of the link)
state (nn) = 2
END DO
state (ic) = 3 ! ic is "dead"
DO WHILE (nper > 0)
i = 1 ! pick a bead of "perimeter" to be analyzed
lab = perlab (i)
ref = perref (i)
perlab (i) = perlab (nper)
perref (i) = perref (nper)
nper = nper - 1
IF (state (lab) == 3) THEN
CYCLE
END IF
state (lab) = 1 ! "lab" is added to the cluster
nclu = nclu + 1
count = count + 1
visit (count) = lab
from (count) = ref
DO k = firstnn (lab), lastnn (lab) ! check nn of newly added
nn = labnn (k)
IF( (state (nn) == 2) .OR. (state (nn) == 3)) CYCLE
nper = nper + 1
perlab (nper) = nn !new bead in perimeter
perref (nper) = lab !its reference bead (origin of the link)
state (nn) = 2
END DO
state (lab) = 3
END DO
nchist (nclu) = nchist (nclu) +1
ic = ic + nclu - 1 ! prepare ic for the next cluster
END DO
WRITE (*,*) "nchist: ", nchist
!-----------------------------------------------------------------------
DEALLOCATE (firstnn, lastnn, deg, labnn)
DEALLOCATE (state)
DEALLOCATE (perlab, perref)
DEALLOCATE (nchist)
RETURN
!-----------------------------------------------------------------------
CONTAINS
!-----------------------------------------------------------------------
SUBROUTINE organize (N, NL, labnn, firstnn, lastnn, deg)
!**********************************************************************
! Analyzes the bonds (bndtbl) to obtain the degree (=number of bonds)
! of each bead, and the nearest neighbours list.
! N in the number of beads (vertices) and NL of bonds (links).
!
! author: s. chiacchiera, February 2017
!**********************************************************************
IMPLICIT none
INTEGER, INTENT(IN) :: N, NL
INTEGER :: i,l,count_lab, i1,i2
INTEGER, DIMENSION (N), INTENT(OUT) :: deg
INTEGER, DIMENSION (N), INTENT(OUT) :: firstnn, lastnn
INTEGER, DIMENSION (2*NL), intent(OUT) :: labnn
deg(:)=0
firstnn(:)=0
lastnn(:)=0
labnn(:)=0
count_lab=0
DO i=1,N
DO l=1,NL
IF(bndtbl(l,1).EQ.i)THEN
deg(i)=deg(i)+1
count_lab=count_lab+1
labnn(count_lab)=bndtbl(l,2)
ENDIF
IF(bndtbl(l,2).EQ.i)THEN
deg(i)=deg(i)+1
count_lab=count_lab+1
labnn(count_lab)=bndtbl(l,1)
ENDIF
END DO
END DO
i1=1
i2=0
DO i=1,N
IF (deg (i)==0) CYCLE
firstnn(i)=i1
i2=i1+deg(i)-1
lastnn(i)=i2
i1=i2+1
END DO
RETURN
END SUBROUTINE organize
!-----------------------------------------------------------------------
END SUBROUTINE connect
|
| [1] | A small change to specifying charge smearing schemes and lengths in CONTROL
files has been made since version 2.6: the old-v2.6 folder includes
CONTROL files for the tests shown here that will work with this version
of DL_MESO. |
| [2] | Disambiguation on the concept of molecule. In DL_MESO a defined molecule is a set of beads, which can be bonded or not. For the purpose of this module it is required that each molecule is a connected cluster (via stretching bonds). In fact, this - together with the reasonable assumption that each stretching bond cannot be stretched to more than half the system linear size - allows us to univocally define the charge dipole moment of each molecule. |
| [3] | The tiny value for 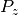 in dipole_BRANCH may vary, but for this test
it should be no greater than the smallest available non-negligible
floating-point number. in dipole_BRANCH may vary, but for this test
it should be no greater than the smallest available non-negligible
floating-point number. |