E-CAM openmm_plectoneme module¶
The openmm_plectoneme is a module that introduces twist rigidity to a polymer and samples the accessible conformations under torsional constraints. This module takes advantage of the OpenMM software and GPU acceleration to perform simulation at the scale of the DNA helix. It builds a Kremer-Grest polymer model with virtual sites to attach a frame to each bead. The frames are used to describe the contour of the molecule and to introduce bending and twisting forces.
Purpose of Module¶
Bacterial DNA is known to form specific conformations called plectonemes because of internal twisting constraints. This physical mechanism participates in the compaction of the genome. The plectonemes are braided structures you often compare with phone cables. In order to study such a system we need to introduce a linking number deficit into a circular polymer.
The Linking number (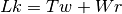 ) is the sum of the twist (
) is the sum of the twist (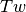 , cumulative
helicity of the DNA) and the writhe (
, cumulative
helicity of the DNA) and the writhe (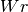 , global intracity).
In the case of circular DNA that is topologically constrained any variation of the
twist affects the Writhe and therefore the conformation.
, global intracity).
In the case of circular DNA that is topologically constrained any variation of the
twist affects the Writhe and therefore the conformation.
In particular, does a slow change of the twist lead to the same conformation that the one we get from a rapidly change in the twist? We then tackle the question : does the introduction protocol of Linking number inside a circular molecule matter? Indeed, does a rapidly Linking number injection freeze the conformation in braided structures where plectonemes do not merge/move along the DNA ? Does the memory of initial conformation matter ?
We can use this module to model single-molecule DNA under magnetic or optical tweezers too. In this kind of setup the molecule is clamped on a plate and to a magnetic bead at the other extremity. The bead is used to apply stretching force and/or rotational constraint. The position of the bead is used to monitor the response of the molecule to the mechanical constraints. From the mechanical constraints you can extract the mechanical properties of your molecule of interest.
This module assist the creation of polymer described by FENE bond and WCA repulsive potential to resolve the excluded volume constraints.
On top of that, the module introduces the twist and mechanical response to twisting constraint with the help of virtual sites functionalities from OpenMM API. The module proposes functions to help the data analysis with High-Performance-Computing Dask software and Python module Numba.
For example, the estimation of the Writhe that is a computation over all the possible pairwise of bonds is highly expensive and can be fasten. In addition to that, we introduce an algorithm to detect the positions, length and shape of plectonemes. It is useful to follow the dynamics of these braided structures and try to answer the previous questions.
This module can be used by polymer physicist to understand the conformation of bacterial DNA under torsional constraints for example. Indeed, it used in a scientific collaboration with Ivan Junier from TIMC-IMAG, Grenoble, France and Ralf Everaers, ENS Lyon, France. However, the publication is not currently available.
Background Information¶
We use the OpenMM toolkit for molecular dynamics. We implemented functionalities to build a frame (that follows the contour of the polymer) and add twisting energy to a Kremer-Grest polymer system. We implemented function to extract plectonemes, writhe and twist from polymer conformations.
Building and Testing¶
The instructions to install, test and run the module can be find on the openmm_plectoneme GitLab repository. The test of the twist implementation can be find at the same location.
We are currently working on a benchmark between the present module and already published Monte-Carlo and rigid body dynamics codes.
Source Code¶
The source code and more information can be find on the openmm_plectoneme GitLab repository.